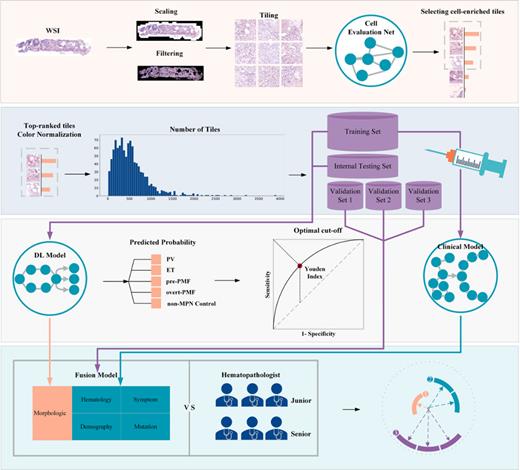
Bone marrow histology was the indispensable tool for the differential diagnosis of classic myeloproliferative neoplasms (MPNs) and subtypes. However, the subjectivity of morphological assessment and markedly overlapping pathological features of different subtypes made accurate diagnosis challenging and controversial. In this study, we developed Clinical, deep learning (DL) and Fusion diagnosis models based on clinical parameters, whole slide images (WSI) based deep learning algorithm using hematoxylin-eosin (HE) staining bone marrow specimen and combination of both for the diagnosis and differentiation of MPNs (Figure 1).
1,051 MPN patients from seven medical centres were enrolled in this study and divided into training, internal testing, internal validation and two external validation cohorts (called combined validation cohort totally). In combined validation cohort, Fusion model performed best in distinguishing MPNs with non-MPN controls with the AUC 0.931 (95%CI: 0.891-0.971). For PV identification, Clinical model achieved the highest AUC with 0.975 (95%CI: 0.960-0.991). Fusion model made best performance in the identification of ET and prePMF, with the AUC 0.887 (95%CI: 0.850-0.925) for ET and 0.899 (95%CI: 0.851-0.947) for prePMF. Misclassified prePMF cases into ET group reduced from 26 (60.5%) in Clinical model to 5 (11.6%) in Fusion model. Consistently, the number of ET cases (N=70, 95.9%) who were misclassified into prePMF in Clinical model reduced to 4 (5.5%) in Fusion model. These results indicated that our Fusion model may have clinical utility in assisting to identify ET and prePMF. Moreover, Fusion model could distinguish overt PMF effectively with AUC 0.980 (95%CI: 0.961-0.999) even with prePMF, and only 3 (7.5%) prePMF cases misclassified into PMF group, suggesting that our machine learning model had high sensitivity in feature identification and extraction.
Next, we compared the performances of the deep learning models with three junior hematopathologists with less than five years of clinical experience and three senior hematopathologists with more than 10 years of experience. 20 cases for each subtype and 20 non-MPN controls, in total 100 cases, were randomly selected from the pool of validation sets with truth label blinded. All the hematopathologists reviewed data and image of 100 patients independently, in parallel with model implementation. Clinical model achieved the highest AUC with 0.925 (0.843-1.000) for PV, which was equivalent with senior hematopathologists (0.929, 0.878-0.979) (difference, 0.004, P=0.8500), while higher than junior ones (0.850, 95%CI: 0.787-0.913) (difference, -0.075, P=0.0007). Fusion model (for ET, 0.806, 95%CI: 0.700-0.913; for prePMF, 0.860, 95%CI: 0.741-0.979) performed better than junior hematopathologists in ET and prePMF identification (for ET, 0.707, 95%CI: 0.539-0.876, P=0.0720; for prePMF, 0.694, 95%CI: 0.564-0.825, P=0.0203), and comparable with senior ones in prePMF and ET identification (for prePMF, 0.787, 95%CI: 0.591-0.984, P=0.2190; for ET, 0.877, 95%CI: 0.860-0.896, P=0.1719). In overt PMF diagnosis, Fusion model (0.952, 95%CI: 0.898-1.000) tended to achieve better performance than both junior (0.850, 95%: 0.774-0.926, P=0.1202) and senior observers (0.823, 95%CI: 0.581-1.000, P=0.0608). The effect sizes could inform future study design for validation.
In conclusion, we developed and externally validated the deep learning models for MPNs diagnosis and subtype differentiation achieving the performances equivalent with senior hematopathologists and better than junior ones. Prospective validation and tool development were underwent to promote the accessibility and feasibility of the proposed models in clinical practice.
Disclosures
No relevant conflicts of interest to declare.